CRISPR/Cas9, TALEN, ZFN system are three powerful genome engineering tools that are composed of programmable, sequence-specific DNA-binding modules associated with a nonspecific DNA nuclease, which introduce DSB (DNA double-strand breaks) that stimulate NHEJ (non-homologous end joining) or HR (homologous recombination) repairing mechanisms at specific genomic locations. In the application of these technology in Drosophila, random indels or mutations can be introduced into DSB via NHEJ, while precise sequence modification can be achieved by providing a proper donor DNA with homologous arms via homologous-directed repair (HDR).
Currently, CRISPR/Cas9 has grown in popularity for Drosophila genome editing.
Drosophila CRISPR/Cas9 System
CRISPR/Cas system (clustered regularly interspaced palindromic repeats/CRISPR associated) is a component of bacteria/archaea innate immunity system. Type II CRISPR/Cas9 system uses ~20 nt sequence of crRNA to bind its complementary sequence of exogenous DNA. Subsequently with the aid of trans-acting RNA (trancrRNA), Cas9 endonuclease is recruited to the sequence and cut off the DNA at both strands.
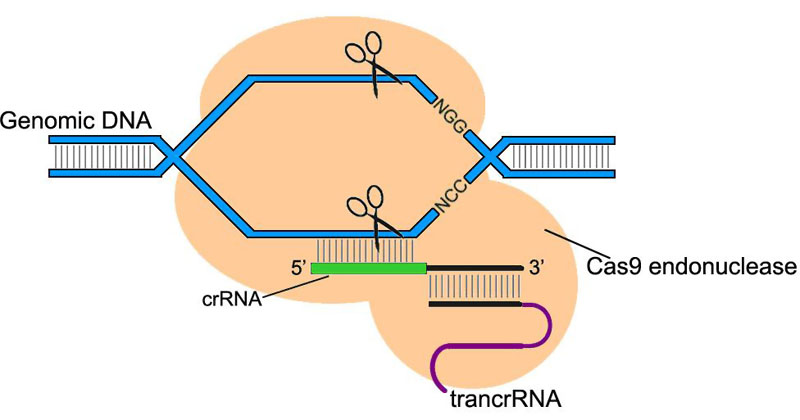
In the application of CRISPR/Cas9 system in Drosophila genome editing, the two RNAs that play key roles can be constructed into a single chimeric RNA (chiRNA or gRNA). By editing the 20 nt targeting sequence of gRNA, the system introduces DSB on the target sequence followed by an NGG or NAG protospacer adjacent motif (PAM) required for Cas9 cleavage.
Since CRISPR/Cas9 works through RNA-DNA recognition, Cas9 can perform its function simultaneously with multiple gRNA target sites, thus facilitates precise deletion, insertion and sequence replacement between the space via HR repair mechanism or multiple sites editing.
Below are three main Drosophila CRISPR/Cas9 systems, please visit their websites for more info:
flyCRISPRFor selecting/designing CRISPR target, Find CRISPRs (flyrnai.org) is a genome-wide resource.
Considering polymorphisms between a chosen fly strain and the reference genome, flyCRISPR Optimal Target Finder identifies gRNA target sites in user-supplied DNA sequence, evaluated with the reference of genome sequences, including D. melanogaster release 6, vas-Cas9(III) (BDSC#51324), nos-Cas9 on II and III
(BDSC#78781 and BDSC#78782) whole-genome sequence, etc.
Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, and O'Connor-Giles KM (2013) Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029-1035
Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, and O'Connor-Giles KM (2013) Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029-1035
Bassett AR, Tibbit C, Ponting CP, Liu JL (2013) Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep 4(1): 220
Kondo S, Ueda R (2013) Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195(3): 715Port F, Chen HM, Lee T, Bullock SL (2014) Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. PNAS 111(29): E2967
Port F, Muschalik N, Bullock SL (2015) Systematic evaluation of Drosophila CRISPR tools reveals safe and robust alternatives to autonomous gene drives in basic research. G3 5(7): 1493
Ren
XJ, Sun J, Housden BE, Hu YH, Roesel C, Lin SL, Liu LP, Yang ZH, Mao
DC, Sun LZ, Wu QJ, Ji JY, Xi JZ, Mohr SE, Xu J, Perrimon N, Ni JQ (2013) Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. PNAS 110(47): 19012
Ren X, Yang Z, Xu J, Sun J, Mao D, Hu Y, Yang SJ, Qiao HH, Wang X, Hu Q, Deng P, Liu LP, Ji JY, Li JB, Ni JQ (2014) Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep 9(3): 1151
Zhang X, Koolhaas WH, Schnorrer F. (2014) A Versatile Two-Step CRISPR- and RMCE-Based Strategy for Efficient Genome Engineering in Drosophila. G3 4(12): 2409
CRISPR/Cas9 Stocks
flyCRISPR Stocks
| Name |
Stock Number
|
Genotype | Description |
Remark
|
| vas-Cas9(X) | BDSC#51323 | y[1] M{vas-Cas9}ZH-2A w[1118] | Expresses Cas9 protein under control of vas regulatory sequences. Construct marked with 3xP3-GFP and attP landing site is marked with 3xP3-RFP | RFP+ GFP+ |
| vas-Cas9(X) RFP- | BDSC#55821 | y[1] M{vas-Cas9.RFP-}ZH-2A w[1118] | The same construct as 51323/51324 transformed to BDSC#24480NF - M{vas-int.Dm}ZH-102D removed and loxP-3xP3-RFP removed version | GFP+ RFP- |
| vas-Cas9(II) | BDSC#56552 | w[1118]; PBac{y[+mDint2]=vas-Cas9}VK00037/CyO, P{w[+mC]=Tb[1]}Cpr[CyO-A] | Expresses Cas9 protein under control of vas regulatory sequences. Construct marked with 3xP3-GFP. w+ linked with CyO | y+ GFP+ w+ CyO |
| vas-Cas9(III) | BDSC#51324 | w[1118]; PBac{y[+mDint2]=vas-Cas9}VK00027 | Expresses Cas9 protein in the ovary under control of vas regulatory sequences. Construct marked with 3xP3-GFP. w[1118]; PBac{y[+mDint2]=vas-Cas9}VK00027/TM6B, Tb[1] version is also available | y+ GFP+ |
FlyCas9 Stocks
| Name | Stock Number | Genotype | Description | Remark |
| nos-Cas9(X) | NIG-FLY#CAS-0002 | y2 cho2 v1 P{nos-Cas9, y+, v+}1A/FM7c, KrGAL4 UAS-GFP | Expresses Cas9 protein under control of nos regulatory sequences. Inserted at the X chromosome | y+ v+ (red eye) |
| nos-Cas9(II-attP40) | NIG-FLY#CAS-0001 | y2 cho2 v1; attP40{nos-Cas9}/CyO | Expresses Cas9 protein under control of nos regulatory sequences. Inserted at the II chromosome, the attP40 site | y+ v+ (v+ hard to see, red eye) |
| yw; nos-Cas9(II-attP40) | - | y1 w1118; attP40{nos-Cas9}/CyO | yw background (much healthier), expresses Cas9 protein under control of nos regulatory sequences. Inserted at the II chromosome, the attP40 site. | y+ CyO (white eye) |
| nos-Cas9(II-2A) | NIG-FLY#CAS-0004 | y2 cho2 v1; Sp/CyO, P{nos-Cas9, y+, v+}2A | Expresses Cas9 protein under control of nos regulatory sequences. Inserted at the II CyO balancer chromosome | y+ v+ Sp CyO (v+ hard to see, red eye) |
| yw; nos-Cas9(III-attP2) | - | y1 w1118; attP2{nos-Cas9}/TM6C,Sb Tb | yw background (much healthier), expresses Cas9 protein under control of nos regulatory sequences. Inserted at the III chromosome, the attP2 site | y+ (white eye) |
| nos-Cas9(III-3A) | NIG-FLY#CAS-0003 | y2 cho2 v1; P{nos-Cas9, y+, v+}3A/TM6C, Sb Tb | Expresses Cas9 protein under control of nos regulatory sequences. Inserted at the III chromosome | y+ v+ TM6C Sb Tb (v+ hard to see, red eye) |
CRISPR fly design Stocks*
| Name | Stock Number | Genotype | Description | Remark |
| Act5C-Cas9.P aka CFD1 | BDSC#54590 | y[1] M{w[+mC]=Act5C-Cas9.P}ZH-2A w[*] | Expresses Cas9 protein ubiquitously under control of Act5C regulatory sequences. Insertion site: ZH-2A(2A3) | w+ RFP+ |
| nos-Cas9.P aka CFD2 | BDSC#54591 | y[1] M{w[+mC]=nos-Cas9.P}ZH-2A w[*] | Expresses Cas9 protein under control of nanos regulatory sequences. Insertion site: ZH-2A(2A3) | w+ RFP+ |
| UAS-Cas9.P aka CFD4 | BDSC#54594 | P{ry[+t7.2]=hsFLP}1, y[1] w[1118]; P{y[+t7.7] w[+mC]=UAS-Cas9.P}attP40 | Expresses Cas9 protein under UAS control. The identity of the hsFLP insertion is a guess. Insertion site: attP40(25C6) | y+ w+ |
| UAS-Cas9.P aka CFD3 | BDSC#54592 | P{ry[+t7.2]=hsFLP}1, y[1] w[1118]; P{y[+t7.7] w[+mC]=UAS-Cas9.P}attP2/TM6B, Tb[1] | Expresses Cas9 protein under UAS control. The identity of the hsFLP insertion is a guess. Insertion site: attP2(68A4) | y+ w+ TM6B Tb |
| nos-Gal4/UAS-Cas9.P aka CFD3-nos | BDSC#54593 | P{ry[+t7.2]=hsFLP}1, y[1] w[1118]; P{y[+t7.7] w[+mC=UAS-Cas9.P}attP2 P{w[+mC]=GAL4::VP16-nos.UTR}CG6325[MVD1] | Expresses Cas9 protein under UAS control. Expresses GAL4 during oogenesis. The identities of the nos-GAL4 and hsFLP insertions are guesses. Breakpoints or Insertion site: attP2(68A4), 86B4 | y+ w+ |
| act-Cas9(D10A) (nickase) aka CFD13 | - | P{ry[+t7.2]=hsFLP}1, y[1] w[1118]; P{y[+t7.7] w[+mC]=act-Cas9D10A}attP2 | Expresses a Cas9 mutant D10A becoming a nickase ubiquitously under control of Act5C regulatory sequences. Insertion site: attP2(68A4) | y+ w+ |
*We only accept Plan RH or Plan RI orders for the Cas9 stocks listed in this table.
nos-Cas9 Stocks*
Name |
Stock Number | Genotype | Description | Remark |
| TH_attP40 nos-Cas9 | BDSC#78781 | y[1] sc[*] v[1] sev[21];{nos-Cas9}attP40/CyO | Expresses Cas9 protein under control of nos regulatory sequences. Inserted at the II chromosome, the attP40 site | y+ v+ CyO |
| TH_attP2 nos-Cas9 | BDSC#78782 | y[1] sc[*] v[1] sev[21];{nos-Cas9}attP2 | Expresses Cas9 protein under control of nos regulatory sequences. Inserted at the III chromosome, the attP2 site | y+ v+ |
Act5C-Cas9, Lig4 Stock
| Name | Stock Number | Genotype | Description | Remark |
| Act5C-Cas9, Lig4 (X) | BDSC#58492 | y[1] M{Act5C-Cas9.P.RFP-}ZH-2A w[1118] Lig4[169] | M{Act5C-Cas9.P}ZH-2A from which miniwhite and 3xP3-RFP were removed by cre recombinase. Act5C-Cas9 on X under lig4169 background | RFP- |
Creating stable gRNA-expressing lines
For creating stable attP-gRNA-expressing lines, you may choose the vermillion+ based attP site on autosomal chromosome lines from PhiC31 lists. We may setup the crosses to the y-v- line and screen for v+. You may get the same v+ G2 transformants as using the v- based attP lines. If v+ transformant on X chromosome, please choose X chromosomal v- background attP line, such as BDSC#34769. This is just regular Plan H or Plan I with extra vermilion+ screening. You may also ask us to keep the resulting gRNA lines and have us cross to Cas9 stock and inject their F1s (additional Plan RG/RH/RI) later.

